A ready-to-use responsive website demo for Team or Academic Lab.
✨ 一个基于 Vue3 + Vite 的静态网页模板,适用于实验室、科研团队、开源项目主页搭建。支持 GitHub Pages 一键部署,实现开盘即用。
🌐 官方示例,TeamWeb preview:https://zaakzoeng.github.io/TeamWeb/ 🌐 中文版使用指南:https://www.yuque.com/parallellight/biometa/tb8mrspti6wggyd2
| Communities | BioMeta, 2BF-Club, AIxOmics |
|---|---|
| Labs | ChenLab |
- 🌐 响应式页面,适配手机/PC 设备
- 🔄 全组件化设计,JSON/图片配置即可更新内容
- 🔗 支持 BibTeX 文献显示和主题/合作单位配置
- 🔧 轻量度和开发者亲和,支持明/暗模式
- ✨ GitHub Pages 直接部署,无需后端
Vite
Vue3
TypeScript
Element Plus
Font Awesome+---vue.config.js // webpack配置
|
+---public
| | favicon.ico // ico图标
| |
| +---assets // 图文资产
| | +---00.basic
| | | background.png // 页面背景图片
| | | logo.svg // 导航栏左侧的logo
| | | parallelLightLogo.png // 底栏Footer左侧的Logo
| | |
| | +---01.home
| | +---02.research
| | +---03.team
| | +---04.publications
| | | | papers.bib // 发表论文的bibtex配置项
| | | |
| | | +---pdf // 发表论文的pdf文件
| | | |
| | | \---preview // 发表论文的preview图片
| | |
| | +---05.resources
| | +---06.moments
| | \---07.contact
| | map.html // 一个Leaflet地图定位HTML文件
| |
| \---jsons // Json配置项
| 00.basic.json // 基本配置项
| 01.home.json // Home首页配置项
| 02.research.json // Research页配置项
| 03.team.json // Team页配置项
| 04.publications.json // Publications页配置项
| 05.resources.json // Resources页配置项
| 06.moments.json // Moments页配置项
| 07.contact.json // Contact页配置项
|
\---src
| App.vue // 根组件
| main.ts // 程序入口文件
|
+---assets
| base.css // 基础样式文件
| main.css
|
+---components
| Backtop.vue // 点击回到顶部组件
| Progress.vue // 进度条组件
|
+---layout
| Footer.vue // 底栏布局
| Header.vue // 导航栏布局
|
+---router
| index.ts // 单页面路由注册组件
|
+---styles
| | index.scss // 基础样式
| |
| \---element
| dark.scss // 暗黑模式
| light.scss // 明亮模式
|
\---views
Contact.vue // 联系我们
Home.vue // 首页
Moments.vue // 时间记录
Publications.vue // 发表刊物
Research.vue // 研究方向
Resources.vue // 工具资源
Team.vue // 团队情况
git clone https://github.com/ZaakZoeng/TeamWeb.git
cd TeamWeb
npm install
npm run dev
# 打开浏览器访问 http://localhost:5173/TeamWeb/详细指南包括:
- 环境准备 (Node.js v22.14.0)
- 克隆项目 (git clone)
- 配置 vite.config.ts 用于 GitHub Pages 部署
- 启动本地服务 npm run dev
- 配置 JSON 和图片替换页面内容
- 构建生产包 npm run build
- 使用 gh-pages 分支部署 GitHub Pages
- Get Emoji:https://getemoji.com/
- Favicon Generator:https://realfavicongenerator.net/
- Contrib Rocks:https://contrib.rocks/
- Star History:https://www.star-history.com/
- Zhong Guo Se:https://www.zhongguose.com/
- 样式整体化,提供用户自行选择修改配色
- 2025.04.18,完善文档
- 2025.04.17 TeamWeb initial commit
下载安装Node.js 配置开发环境(建议选择v22.14.0版本)
验证Node.js和npm是否安装成功,出现版本号即成功
> node -v
v22.14.0
> npm -v
10.9.2从Github中克隆该项目到本地。
git clone git@github.com:ZaakZoeng/TeamWeb.git
# git clone your repo也可以直接点击“Use this template”选择“Create a new repository”,自定义命名(⚠⚠⚠请注意自定义命名时和后面使用GitHub Pages部署息息相关,如果你计划使用https://yourname.github.io/TeamWeb/作为域名,则直接按照本指南操作即可;如果你计划直接使用https://yourname.github.io/作为域名,则需要克隆后在vite.config.ts中修改代码)。
vite.config.ts
import { defineConfig } from 'vite';
export default defineConfig({
base: "./", // 设置部署路径为 ./,使用 GitHub Pages 时对应https://yourname.github.io/,
// base: "/TeamWeb/", // 设置部署路径为 /TeamWeb/,使用 GitHub Pages 时对应https://yourname.github.io/TeamWeb/
});进入项目目录,并安装项目对应的依赖包
> cd TeamWeb
> npm install运行项目进入开发模式,浏览器访问本地地址:http://localhost:5173/TeamWeb/ 可预览项目(此时项目已经在本地 run 起来了,在开发模式中继续配置数据可实时刷新预览结果)
> npm run dev
VITE v4.5.11 ready in 696 ms
➜ Local: http://localhost:5173/TeamWeb/
➜ Network: http://172.29.172.208:5173/TeamWeb/
➜ press h to show helpTeamWeb 是基于页面独立的逻辑进行开发完成的,即每一个页面使用一个独立的配置文件,用户可以开箱即用,直接更改/public/assets中的资产内容,然后修改/public/jsons中的配置项对数据内容更新。
基础配置主要是针对 Header 导航栏和 Footer 底栏的图文配置,用户可以更换为自己的内容。
/public/jsons/00.basic.json
{
"header": {
"logo": "/assets/00.basic/logo.svg" // 导航栏左侧Logo,图片链接或图片文件路径
},
"footer": {
"logo": "/assets/00.basic/parallelLightLogo.png", // 底栏左侧Logo,图片链接或图片文件路径
"logoLink": "",
"clustrmaps": "//clustrmaps.com/map_v2.png?cl=ffffff&w=100&t=tt&d=tn5kfLlfmc04L5tfIH3RXxlb4N3Hd7hZmmq0gAAnMZs", // 底栏中间的clustrmaps配置项,可访问https://clustrmaps.com/生成并替换
"poweredList": [ // 底栏右侧的开发者信息,可添加多个name和link
{
"name": "Ze Zhang",
"link": "https://zaakzoeng.github.io/"
}
]
}
}首页主要展示了实验室主题以及 Hot News 内容。
/public/jsons/01.home.json
{
"basic": {
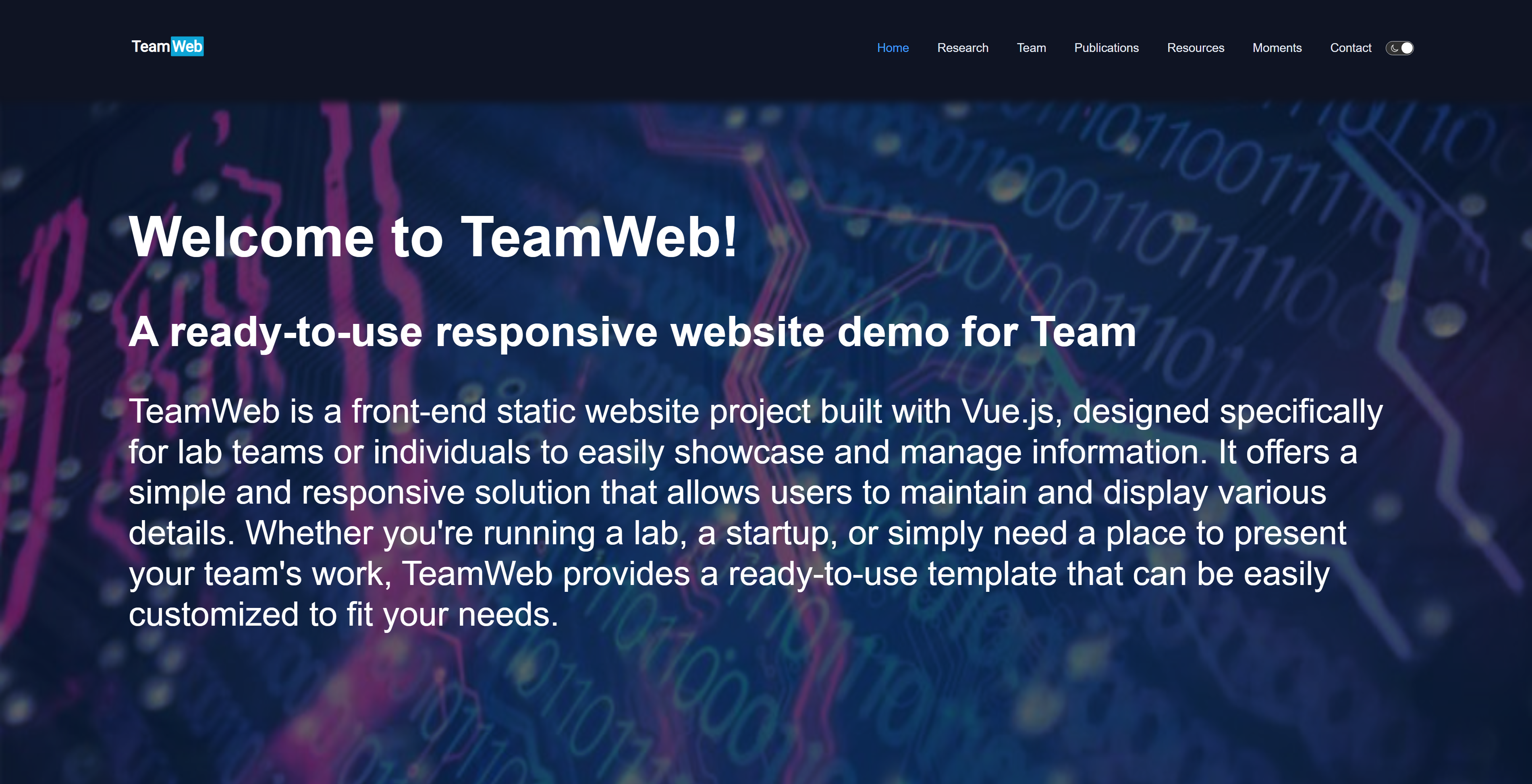
"title": "Welcome to TeamWeb!",
"subtitle": "A ready-to-use responsive website demo for Team",
"introduction": "TeamWeb is a front-end static website project built with Vue.js, designed specifically for lab teams or individuals to easily showcase and manage information. It offers a simple and responsive solution that allows users to maintain and display various details. Whether you're running a lab, a startup, or simply need a place to present your team's work, TeamWeb provides a ready-to-use template that can be easily customized to fit your needs."
}, // Home首页基础配置项信息
"news": [ // 首页Hot News信息
{
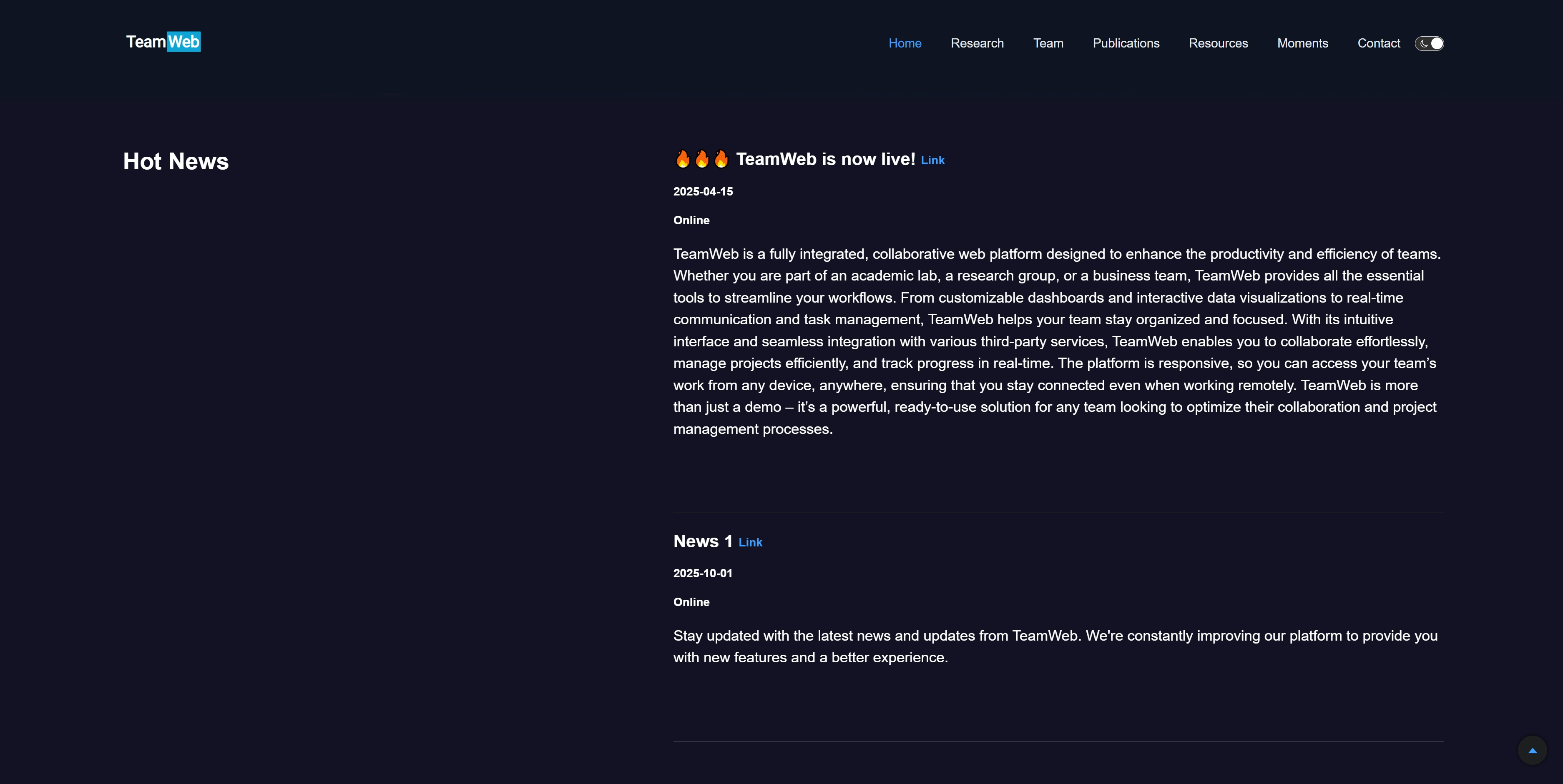
"title": "🔥🔥🔥 TeamWeb is now live!", // 文本
"link": "https://zaakzoeng.github.io/TeamWeb/", // 链接
"datetime": "2025-04-15", // 时间格式XXXX-XX-XX
"address": "Online", // 文本
"introduction": "TeamWeb is a fully integrated, collaborative web platform designed to enhance the productivity and efficiency of teams. Whether you are part of an academic lab, a research group, or a business team, TeamWeb provides all the essential tools to streamline your workflows. From customizable dashboards and interactive data visualizations to real-time communication and task management, TeamWeb helps your team stay organized and focused. With its intuitive interface and seamless integration with various third-party services, TeamWeb enables you to collaborate effortlessly, manage projects efficiently, and track progress in real-time. The platform is responsive, so you can access your team’s work from any device, anywhere, ensuring that you stay connected even when working remotely. TeamWeb is more than just a demo – it’s a powerful, ready-to-use solution for any team looking to optimize their collaboration and project management processes.", // 文本
"isShow": true // 可修改isShow来决定是否展示信息
},
{
"title": "News 1",
"link": "https://zaakzoeng.github.io/TeamWeb/",
"datetime": "2025-10-01",
"address": "Online",
"introduction": "Stay updated with the latest news and updates from TeamWeb. We're constantly improving our platform to provide you with new features and a better experience.",
"isShow": true
}
]
}Research 页主要展示团队的多个研究方向介绍。
/public/jsons/02.research.json
{
"basic": {}, // Research页基础配置项信息
"researches": [
{
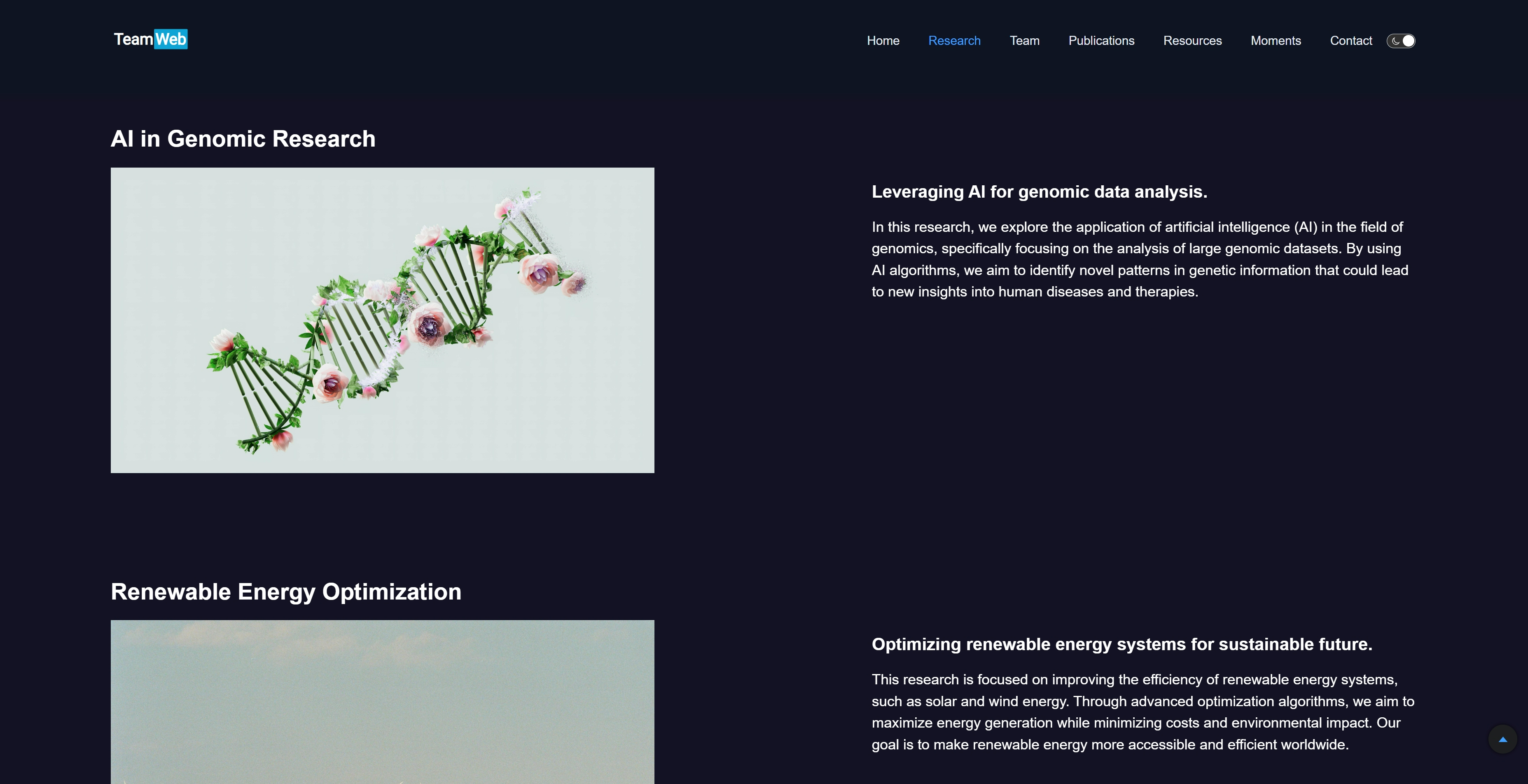
"title": "AI in Genomic Research",
"image": "/assets/02.research/Research-AI.jpg",
"subtitle": "Leveraging AI for genomic data analysis.",
"content": "In this research, we explore the application of artificial intelligence (AI) in the field of genomics, specifically focusing on the analysis of large genomic datasets. By using AI algorithms, we aim to identify novel patterns in genetic information that could lead to new insights into human diseases and therapies.",
"isShow": true
},
{
"title": "Renewable Energy Optimization",
"image": "/assets/02.research/Research-RenewableEnergy.jpg",
"subtitle": "Optimizing renewable energy systems for sustainable future.",
"content": "This research is focused on improving the efficiency of renewable energy systems, such as solar and wind energy. Through advanced optimization algorithms, we aim to maximize energy generation while minimizing costs and environmental impact. Our goal is to make renewable energy more accessible and efficient worldwide.",
"isShow": true
}
]
}Team 页展示团队成员、合作单位等信息。
/public/jsons/03.team.json
{
"basic": {}, // Team页基础配置项信息
"persons": [
{
"avatar": "/assets/03.team/team-ZhangZe.jpg", // 头像,方形图片,链接或文件路径
"name": "Ze Zhang", // 姓名
"position": "Ph.D. candidate student", // 职位
"website": "https://zaakzoeng.github.io/", // 个人网页链接
"commerce": [ // commerce中如果有不想展示的内容,则需要将account修改为""空值
{
"icon": [
"fas",
"envelope"
],
"account": "zhangze21@mails.ucas.ac.cn", // 邮箱
"link": "mailto:zhangze21@mails.ucas.ac.cn" // 邮箱
},
{
"icon": [
"fab",
"google"
],
"account": "Ze's Google Scholar", // 谷歌学术
"link": "https://scholar.google.com/citations?user=6eXARMsAAAAJ" // 谷歌学术链接
},
{
"icon": [
"fab",
"github"
],
"account": "Ze's Github", // Github
"link": "https://github.com/ZaakZoeng" // Github链接
},
{
"icon": [
"fab",
"twitter"
],
"account": "", // 推特,此时为空则不会在网页中展示出来,其他同理
"link": "" // 推特链接
},
{
"icon": [
"fab",
"linkedin"
],
"account": "", // 领英
"link": "" // 领英链接
},
{
"icon": [
"fab",
"facebook"
],
"account": "", // 脸书
"link": "" // 脸书链接
},
{
"icon": [
"fab",
"zhihu"
],
"account": "", // 知乎
"link": "" // 知乎链接
},
{
"icon": [
"fas",
"link"
],
"account": "Ze's Personal Website", // 个人网页
"link": "https://zz.biometa.top/" // 个人网页链接
},
{
"icon": [
"fab",
"weixin"
],
"account": "ZaakZoeng" // 微信号
},
{
"icon": [
"fab",
"qq"
],
"account": "2216672515" // QQ号
},
{
"icon": [
"fas",
"phone"
],
"account": "+86-19276868848" // 电话或手机号
}
],
"interests": [ // 研究方向,数组列表格式
"Bioinformatics",
"Database tools",
"Single cell & ST Multi omics"
],
"biography": "Ze received his bachelor's degree in information security from Nanchang university in 2021.", // 个人简介,,如果为空则不展示
"hometown": "Ningjin, Xingtai, Hebei, China", // 家乡,如果为空则不展示
"fact": "He always wanted to be someone who could go to bed early and get up early.", // 座右铭或个人有趣的事儿,如果为空则不展示
"timeline": "2021.06 - Now", // 加入团队时间
"isShow": true // 是否展示该成员信息,如果为false则不展示
},
{},
{},
{}
],
"supports": [ // Team页合作单位的Supports信息
{
"unit": "HIAS",
"logo": "http://hias.ucas.ac.cn/images/t_logo.png",
"link": "http://hias.ucas.ac.cn/"
},
{
"unit": "CAS",
"logo": "https://www.zkszl.net/zhongke/userfiles/1/images/zk-logo.png",
"link": "https://www.cas.cn/"
},
{
"unit": "CEMCS",
"logo": "http://cemcs.cas.cn//images/shxbs_logo.png",
"link": "http://cemcs.cas.cn/"
},
{
"unit": "UCAS",
"logo": "/assets/03.team/supports-UCAS.png",
"link": "https://www.ucas.ac.cn/"
}
]
}Publications 主要展示刊物发表情况。
/public/jsons/04.publications.json
{
"basic": {
"subtitle": "The show of our work",
"slogan": "Access the lab's published research and news media coverage"
}
}值得注意的是,《刊物发表》的配置项与其他页面配置方式有所不同,为了更便于文献的展示,项目使用了常见的bibtex引文格式,用户直接复制文献的 bib 引文到 /public/assets/04.publications/papers.bib 中即可完成配置,除此之外,每个 paper 还支持额外配置“keywords、google_scholar、pmid、website、chinese_report、pdf、citation”等内容。
/public/assets/04.publications/papers.bib
---
---
@article{10.3389/fmicb.2025.1557285,
abbr = {Frontiers in Microbiology},
preview = {/assets/04.publications/preview/20250321-fmicb-FACdb.jpg},
bibtex_show = {true},
html = {https://www.frontiersin.org/journals/microbiology/articles/10.3389/fmicb.2025.1557285/full},
selected = {true},
author = {Zhang†, Ze and Li†, Yang and Zhang†, Di and Chen*, Shuai and Lu, Sien and Wang, Kang and Zhou, Miao and Song, Zehe and Li*, Qingcui and Yin*, Jie and Liu*, Xiaoping },
title = {FACdb: a comprehensive resource for genes, gut microbiota, and metabolites in farm animals},
journal = {Frontiers in Microbiology},
year = {2025},
month = {03},
day = {21},
pages = {},
volume = {16},
number = {},
doi = {10.3389/fmicb.2025.1557285},
url = {https://www.frontiersin.org/journals/microbiology/articles/10.3389/fmicb.2025.1557285/full},
eprint = {https://www.frontiersin.org/journals/microbiology/articles/10.3389/fmicb.2025.1557285/full},
abstract = {Farm animals, including livestock and poultry, play essential economic, social, and cultural roles and are indispensable in human welfare. Farm Animal Connectome database (FACdb) is a comprehensive resource that includes the association networks among gene expression, gut microbiota, and metabolites in farm animals. Although some databases present the relationship between gut microbes, metabolites, and gene expression, these databases are limited to human and mouse species, with limited data for farm animals. In this database, we calculate the associations and summarize the connections among gene expression, gut microbiota, and metabolites in farm animals using six correlation or distance calculation (including Pearson, Spearman, Cosine, Euclidean, Bray–Curtis, and Mahalanobis). FACdb contains over 55 million potential interactions of 73,571 genes, 11,046 gut microbiota, and 4,540 metabolites. It provides an easy-to-use interface for browsing and searching the association information. Additionally, FACdb offers interactive visualization tools to effectively investigate the relationship among the genes, gut microbiota, and metabolites in farm animals. Overall, FACdb is a valuable resource for understanding interactions among gut microbiota, metabolites, and gene expression. It contributes to the further utilization of microbes in animal products and welfare promotion. Compared to mice, pigs or other farm animals share more similarities with humans in molecular, cellular, and organ-level responses, indicating that our database may offer new insights into the relationship among gut microbiota, metabolites, and gene expression in humans.},
keywords = {association networks; connectome database; farm animal; gene expression; gut microbiota; metabolites},
google_scholar = {https://scholar.google.com/scholar?hl=en&as_sdt=0%2C5&q=FACdb%3A+a+comprehensive+resource+for+genes%2C+gut+microbiota%2C+and+metabolites+in+farm+animals&btnG=},
pmid = {40190740},
website = {http://122.224.251.240:2023/FACdb/},
chinese_report = {},
pdf = {/assets/04.publications/pdf/20250321-fmicb-FACdb.pdf},
citation = {Zhang, Z., Li, Y., Zhang, D., Chen, S., Lu, S., Wang, K., Zhou, M., Song, Z., Li, Q., Yin, J., & Liu, X. (2025). FACdb: a comprehensive resource for genes, gut microbiota, and metabolites in farm animals. Frontiers in microbiology, 16, 1557285. https://doi.org/10.3389/fmicb.2025.1557285}
}Resources 页面主要展示各种资源链接,提供一个公共的跳板。
/public/jsons/05.resources.json
{
"basic": {
"subtitle": "The resources of our work",
"slogan": "Obtain codes, database tools, and resources"
},
"resources": [
{
"title": "Database",
"subtitle": "Databases developed by the group",
"tools": [
{
"name": "FACdb",
"desc": "Farm Animal Connectome database",
"link": "http://compbiol.top:2023/FACdb/",
"isShow": true
},
{
"name": "CASA",
"desc": "Cell Atlas involving Sex and Androgens",
"link": "https://casadbtools.com/",
"isShow": true
},
{
"name": "RSIT",
"desc": "Earthquake Alerting System",
"link": "https://earthquakepredictionrsit.com/",
"isShow": true
},
{
"name": "TeCD",
"desc": "The eccDNA Collection Database",
"link": "http://122.224.251.240:2022/",
"isShow": true
}
],
"isShow": true
},
{
"title": "Source Code",
"subtitle": "Source code provided by the group",
"tools": [
{
"name": "SSN4PaCDM",
"desc": "Constructing sample-specific networks (SSN) for different sample types to analyze pancreatic cancer (PaC)-associated diabetes mellitus (PaCDM)",
"link": "https://github.com/ZaakZoeng/SSN4PaCDM",
"isShow": true
}
],
"isShow": true
},
{
"title": "AI Chat Tools",
"subtitle": "Some useful AI chat tools",
"tools": [
{
"name": "Tongyi QWen",
"desc": "Intelligent conversational assistant by Alibaba Cloud",
"link": "https://qianwen.aliyun.com/",
"isShow": true
},
{
"name": "DeepSeek",
"desc": "Large language model developed by DeepSeek Inc.",
"link": "https://www.deepseek.com/",
"isShow": true
},
{
"name": "ChatGPT",
"desc": "Conversational AI system from OpenAI",
"link": "https://chat.openai.com/",
"isShow": true
},
{
"name": "Claude",
"desc": "AI assistant created by Anthropic",
"link": "https://claude.ai/",
"isShow": true
},
{
"name": "Gemini",
"desc": "Multimodal AI model by Google",
"link": "https://gemini.google.com/",
"isShow": true
}
],
"isShow": true
}
]
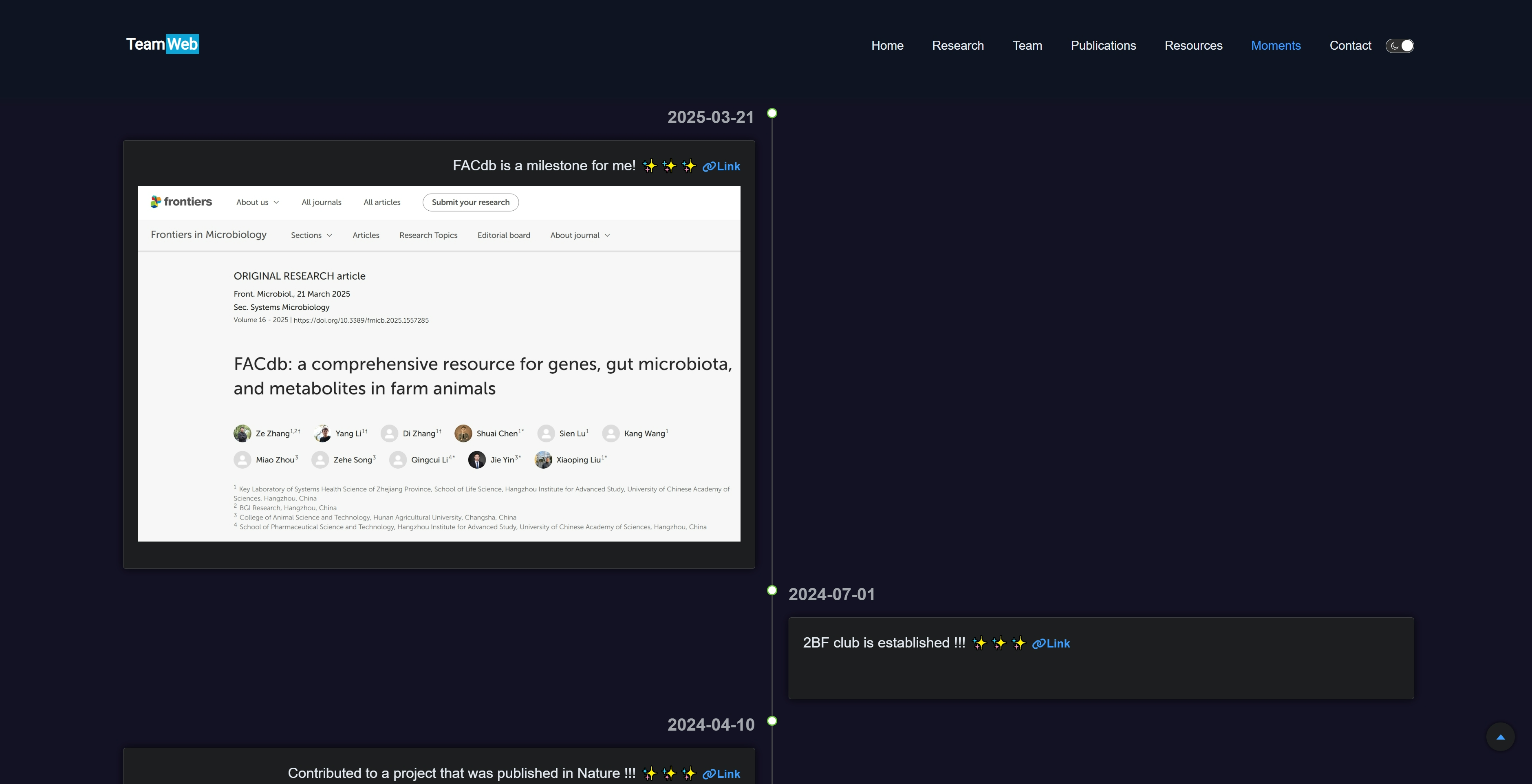
}Moments 页面记录团队的高光时刻。
整个项目支持响应式布局,即在不同大小设备屏幕上有相应的布局展示,在 Moments 页面中尤为明显,具体体现在宽屏时为左右对称布局展示 moments 内容,窄屏时则统一调整至左侧时间线展示 moments。
/public/jsons/06.moments.json
{
"basic": {
"subtitle": "Highlight Moment of Lab",
"slogan": "Browse laboratory happy moments"
},
"moments": [
{
"date": "2025-03-21",
"title": "FACdb is a milestone for me! ✨✨✨",
"link": "https://www.frontiersin.org/journals/microbiology/articles/10.3389/fmicb.2025.1557285/full",
"image": "/assets/06.moments/20250321-FACdb.jpeg",
"isShow": true
},
{
"date": "2024-07-01",
"title": "2BF club is established !!! ✨✨✨",
"link": "https://2bf.biometa.top/",
"image": "",
"isShow": true
},
{
"date": "2024-04-10",
"title": "Contributed to a project that was published in Nature !!! ✨✨✨",
"link": "https://casadbtools.com/",
"image": "/assets/06.moments/20240410-Nature_CASA.png",
"isShow": true
}
]
}Contact 是联系信息的展示。Contact 页面中引入的地图是 Leaflet 地图的 HTML 格式文件,用户可以自行前往 https://leafletjs.com/ 使用定义。
/public/jsons/07.contact.json
{
"basic": {
"subtitle": "Our Detail Information",
"slogan": "Contact us at any time"
},
"content": {
"name": "Ze Zhang",
"institution": "Indie developer",
"address": "Hangzhou 310030, Zhejiang Province, China",
"phone": "+86-19276868848",
"email": "zaakzoeng@gmail.com"
},
"map": "/assets/07.contact/map.html"
}开发完成后需要打包项目,打包完成后会出现/dist文件夹
> npm run build提前下载好 Git 工具。这里仅分享直接使用GitHub Pages部署 TeamWeb 项目,本质上是利用Github Pages部署基于Vue开发的静态页面的方法。为了方便用户的代码能够完整保存与长远维护,本方法采用“主干与分支”的思路进行分别处理:
- 主干 main:用来存储全部代码
- 分支 gh-pages:用来部署静态页面
首先,需要将所有代码使用git指令上传到该项目的主干main中
# 添加要上传的全部文件
> git add .
# 提交更改
> git commit -m "All Codes"
# 从 github 上拉取至本地
> git pull
# 从本地上传至 github
> git push其次,将打包好的dist上传到该项目的分支gh-pages中
# 查看分支
> git branch
# 如果没有 gh-pages 分支,则可以新建并进入分支 gh-pages,
> git checkout -b gh-pages
# 清空 gh-pages 分支的内容
> git rm -rf .
# 将 dist 目录的内容复制到当前目录(即 gh-pages 分支)
> cp -r dist/* .
# 添加要上传的全部文件
> git add .
# 提交更改
> git commit -m "Deploy Vue project to GitHub Pages"
# 如果需要,使用变基
# git config pull.rebase true
# 从 github 中的 gh-pages 分支上拉取至本地
> git pull origin gh-pages
# 将 gh-pages 分支从本地推送到 GitHub
> git push origin gh-pages
# 如果需要,可删除本地
# git branch -d gh-pages然后,在Github的该项目中选择“Settings”,在“General”中选择“Pages”。再选择“Deploy from a branch”,稍等完成处理。
The theme is available as open source under the terms of the MIT License.