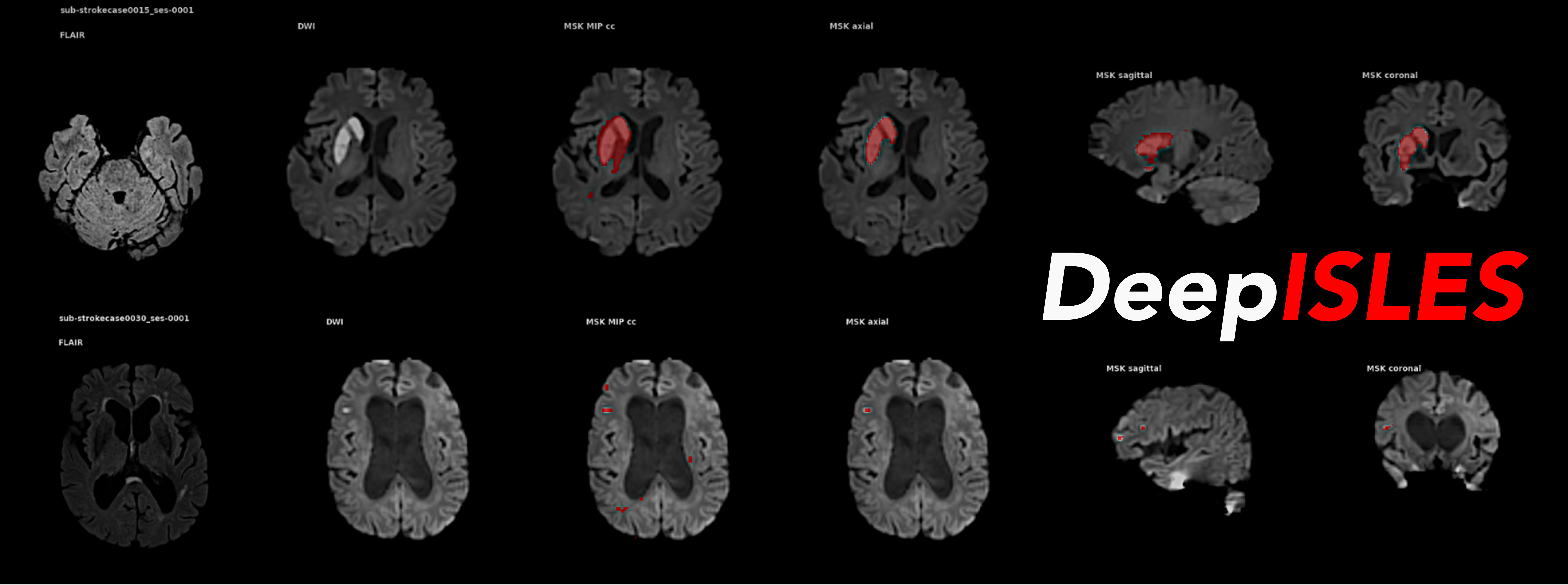
DeepISLES is an out-of-the-box software tool for processing MRI scans and segmenting ischemic stroke lesions, developed in collaboration with leading teams from the ISLES'22 MICCAI Challenge.
Content:
DeepISLES is available in four different formats, catering to various use cases, from an easy-to-use web service to a command-line implementation for batch-processing large data sets. Below, you will find installation and use instructions for the various formats. We provide an example MRI scan from the ISLES'22 dataset (Hernandez Petzsche et al., Sci Data 2022) in /data/:
DeepISLES is available as an easy-to-use web service, hosted on Grand Challenge. This service allows for an easy, straight-forward processing of individual scans to try out DeepISLES.
To access the web service, please follow these three steps:
- Create an account on Grand Challenge (https://grand-challenge.org/) AND verify it. Please note that GC sends out a validation email that contains instructions how to successfully activate your account.
- Request access to DeepISLES ("Try out algorithm").
- Drag-and-drop the MRI scans, wait until the job ends, and download your results!
Note: DICOM inputs must be provided for each MR sequence as a single .zip file for the web service.
A standalone version of DeepISLES is available, complete with a graphical user interface (GUI) that supports both single case and batch processing. This version is hosted on NITRC ("Downloads" section on the left-hand side). Installation instructions for Linux and Windows systems (GPU required) are also provided there.
Please note that the standalone software also requires Docker + Nvidia Container Toolkit installed - see Docker for details!
Note: Ensure your data is organized so that all MR modalities are contained within a single folder for .nii/.nii.gz/.mha files (e.g., /path_to_data/dwi.nii.gz, /path_to_data/adc.nii.gz, /path_to_data/flair.nii.gz) or, for DICOM files, with each modality in separate subfolders (e.g., /path_to_dicom/dwi, /path_to_dicom/adc, /path_to_dicom/flair).
For easy command-line usage, we provide a pre-built docker image.
- Install Docker and NVIDIA Container Toolkit
- IMPORTANT: Verify your docker / Nvidia installation using
docker run --rm --runtime=nvidia --gpus all ubuntu nvidia-smi. This command should output a list of your NVidia GPUs, ensuring that they are accessible in Docker also. - Download the Docker image (
docker pull isleschallenge/deepisles)
docker run --gpus all -v */path_to_data*:/app/data isleschallenge/deepisles --dwi_file_name dwi.nii.gz --adc_file_name adc.nii.gz --flair_file_name flair.nii.gzFor DICOM usage, organize your data directory into subfolders (e.g., /path_to_data/dwi_folder, /path_to_data/adc_folder, /path_to_data/flair_folder), and pass the folder names as arguments by running:
docker run --gpus all -v */path_to_data*:/app/data isleschallenge/deepisles --dwi_file_name dwi_folder --adc_file_name adc_folder --flair_file_name flair_folder Note: Please replace */path_to_data* with the path where you store your image data files, for example, /mnt/media/data.
Note: By default, the docker will save results into a (newly-created) subfolder named results/ in that folder (e.g. /mnt/media/data/results)
skull_strip:True/False(default:False) — Perform skull stripping on input images.fast:True/False(default:False) — Run a single model for faster execution.parallelize:True/False(default:True) — Up to 50% faster inference on GPUs with ≥12 GB memory.save_team_outputs:True/False(default:False) — Save outputs of individual models before ensembling.results_mni:True/False(default:False) — Save images and outputs in MNI.
1.1) Clone this repository.
git clone https://github.com/ezequieldlrosa/DeepIsles.git
cd DeepIsles1.2) Create a conda environment and install dependencies. Note: Mandatory Python version 3.8.0 (!)
conda create --name deepisles python=3.8.0 pip=23.3.1
conda activate deepisles
conda install pytorch==1.11.0 torchvision==0.12.0 torchaudio==0.11.0 cudatoolkit=11.3 -c pytorch
conda install -c conda-forge openslide-python
conda install python=3.8.0 # important, since pytorch triggers the installation of later python versions
pip install -e ./src/SEALS/
pip install -e ./src/FACTORIZER/model/factorizer/
pip install -e ./src/HD-BET
pip install -r requirements.txt
If you successfully installed all required packages, you can follow the steps below to download and place the checkpoints.
1.3) Download the model weights from here and decompress the file inside this repo. From the terminal:
wget https://zenodo.org/records/14026715/files/stroke_ensemble_weights.7z?download=1
mv 'stroke_ensemble_weights.7z?download=1' weights.7z
7za x weights.7z
Your directory should look like this:
DeepIsles/
├── weights/
│ ├── SEALS/
│ │ └── (...)
│ ├── NVAUTO/
│ │ └── (...)
│ └── FACTORIZER/
│ └── (...)
- Input formats:
.dcm,.nii,.nii.gz,.mha. - Processing: The algorithm works directly in the native image space — no additional preprocessing is required.
- DWI (b=1000): Required
- ADC: Required
- FLAIR: Required for ensemble (optional for single algorithm outputs)
PATH_DEEPISLES = 'path-to-repo'
import sys
sys.path.append(PATH_DEEPISLES)
from src.isles22_ensemble import IslesEnsemble
INPUT_FLAIR = 'path-to-flair.nii.gz'
INPUT_ADC = 'path-to-adc.nii.gz'
INPUT_DWI = 'path-to-dwi.nii.gz'
OUTPUT_PATH = 'path-to-output-folder'
stroke_segm = IslesEnsemble()
stroke_segm.predict_ensemble(ensemble_path=PATH_DEEPISLES,
input_dwi_path=INPUT_DWI,
input_adc_path=INPUT_ADC,
input_flair_path=INPUT_FLAIR,
output_path=OUTPUT_PATH,
skull_strip=False,
fast=False,
save_team_outputs=False,
results_mni=False,
parallelize=True
)Try DeepISLES out with the provided example data:
python scripts/predict.pyIf you use this repository, please cite the following publications:
-
de la Rosa, E., Reyes, M., Liew, S. L., Hutton, A., Wiest, R., Kaesmacher, J., ... & Wiestler, B. (2025).
DeepISLES: A clinically validated ischemic stroke segmentation model from the ISLES'22 challenge
Nature Communications -
Hernandez Petzsche, M. R., de la Rosa, E., Hanning, U., Wiest, R., Valenzuela, W., Reyes, M., ... & Kirschke, J. S. (2022).
ISLES 2022: A multi-center magnetic resonance imaging stroke lesion segmentation dataset.
Scientific Data, 9(1), 762
-
Algorithm SWAN is based on FACTORIZER. Git repo
DeepISLES brings together three heterogeneous algorithms, each developed independently by leading research teams around the world. These algorithms rely on distinct libraries, preprocessing protocols, and training methodologies. This diversity is a key factor in DeepISLES’ robust performance, but it also makes direct fine-tuning of the individual models quite challenging in practice. To address this and to support ongoing community-driven development, we here outline two practical fine-tuning strategies:
- We provide comprehensive instructions for incorporating new models into the DeepISLES ensemble:
-
Add your model's inference code
Include your inference script (Python or Bash) inside theinference()function. -
Adapt the ensembling logic
Modify themajority_voting.pyscript to read and ensemble your model’s outputs.
You can choose to include or exclude any of the built-in DeepISLES models (SEALS, NVAUTO, SWAN) depending on your specific application.
- As a more straightforward alternative, we document how to adjust the weighting of the three models implemented in the DeepISLES ensemble:
- Adapt the ensembling logic
Modify themajority_voting.pyscript to adjust how the three models are weighted. Alternatively, you can set thesave_team_outputsflag toTruein the DeepISLES docker and implement a custom logic for fusing the three model results.
Please contact Ezequiel de la Rosa (ezequiel.delarosa@uzh.ch).
- We thank all ISLES'22 challenge participants, collaborators and organizers for allowing this work to happen. We also thank all developers and maintaners of the repos herein used.
- Skull-stripping is done with HD-BET.
- The used FLAIR-MNI atlas is obtained from this paper (https://zenodo.org/records/3379848).