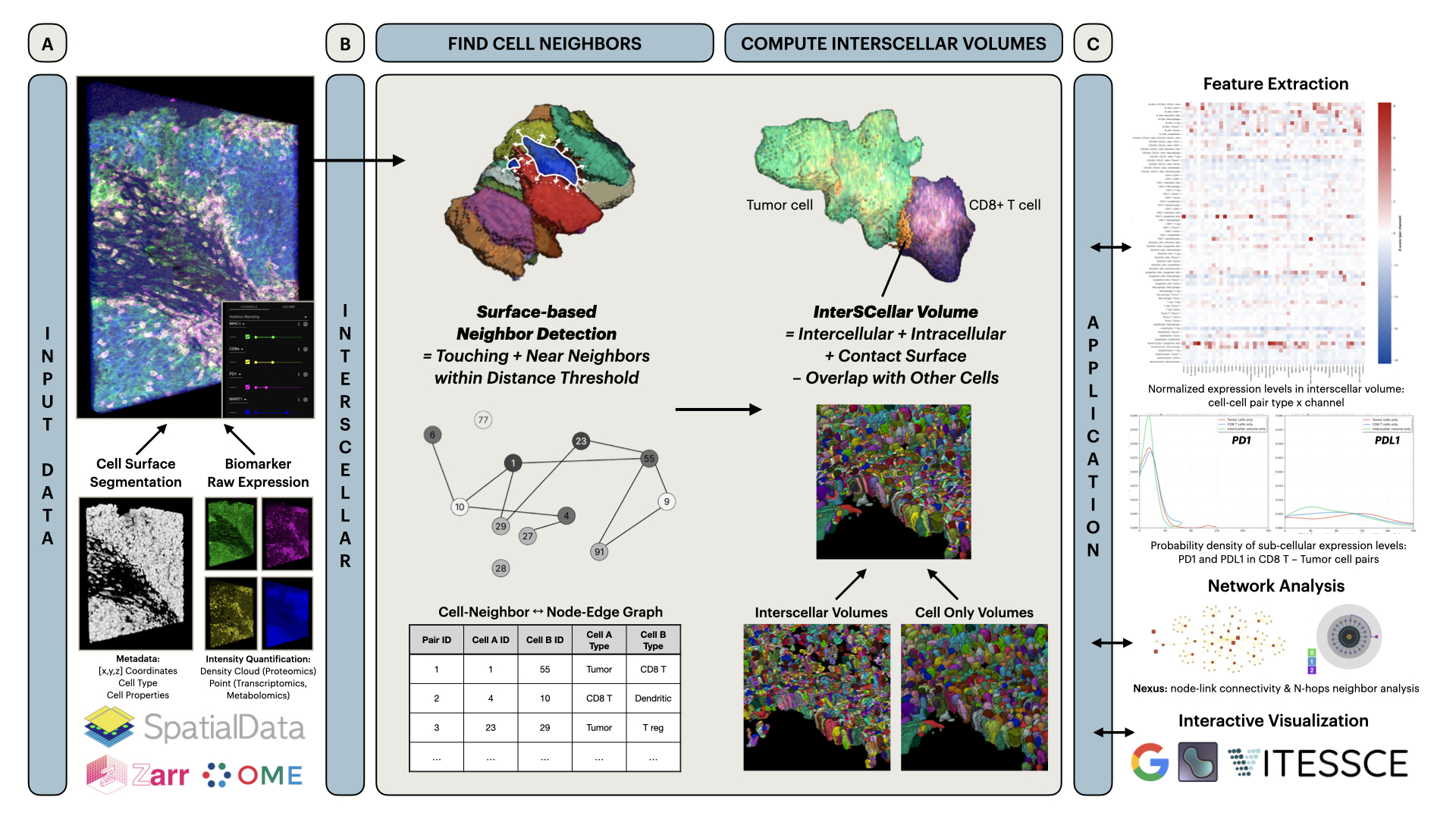
InterSCellar is a Python package for surface-Based cell neighborhood and interaction volume analysis in 3D spatial omics.
Install package:
pip install interscellarImport:
import interscellar(1) Cell Neighbor Detection & Graph Construction
neighbors_3d, adata, conn = interscellar.find_cell_neighbors_3d(
ome_zarr_path="data/segmentation.zarr",
metadata_csv_path="data/cell_metadata.csv",
max_distance_um=0.5,
voxel_size_um=(0.56, 0.28, 0.28),
n_jobs=4
)(2) Interscellar Volume Computation
# Interscellar volumes
volumes_3d, adata, conn = interscellar.compute_interscellar_volumes_3d(
ome_zarr_path="data/segmentation.zarr",
neighbor_pairs_csv="results/neighbors_3d.csv",
neighbor_db_path="/results/neighbor_graph.db",
voxel_size_um=(0.56, 0.28, 0.28),
max_distance_um=3.0,
intracellular_threshold_um=1.0,
n_jobs=4
)# Cell-only volumes
cellonly_3d = interscellar.compute_cell_only_volumes_3d(
ome_zarr_path="data/segmentation.zarr",
interscellar_volumes_zarr="results/interscellar_volumes.zarr"
)(1) Cell Neighbor Detection & Graph Construction
neighbors_2d, adata, conn = interscellar.find_cell_neighbors_2d(
polygon_json_path="data/cell_polygons.json",
metadata_csv_path="data/cell_metadata.csv",
max_distance_um=1.0,
pixel_size_um=0.1085,
n_jobs=4
)Volume Visualization
# Full dataset (Napari)
visualize-all-3d \
--cell-only-zarr "results/cell_only_volumes.zarr" \
--interscellar-zarr "results/interscellar_volumes.zarr" \
--cell-only-opacity 0.7 \
--interscellar-opacity 0.9# Single pair (Napari)
visualize-pair-3d \
--pair-id 123 \
--cell-only-zarr "results/cell_only_volumes.zarr" \
--interscellar-zarr "results/interscellar_volumes.zarr" \
--pair-opacity 0.6 \
--cells-opacity 0.7