-
Notifications
You must be signed in to change notification settings - Fork 0
Home
MITgcm_BFM_chain is the overarching algorithm of the high-resolution, relocatable modeling system for short-term forecasts developed in the framework of the SHAREMED (MED Programme) strategic project. The code has been implemented during the project and capitalizes on the OGS ECHO group experience in hydrodynamic-biogeochemical modeling and operational oceanography.
The relocatable system is based on the coupled hydrodynamic-biogeochemical model MITgcm-BFM, and it is initialized and forced by the Copernicus Marine Environment Monitoring Service (CMEMS-CMS) and other available operational products (e.g., satellite data, in-situ measurements). The forecasting system was designed to be easily relocatable: it was firstly implemented in operational mode in the northern Adriatic study site, then it was further tested and applied in a cluster of transnational coastal areas in the Mediterranean Sea (i.e., Sicily Channel -WP4.5- and North Western Med and sub-basins -WP5.1-). All the local implementations were customized by adapting the python and bash scripts developed for the pre- and post-processing phases of each operational system, which shapes up to be an effective regional downscaling of the Copernicus Marine Service (CMS) Analysis and Forecast products.
MITgcm_BFM_chain has been designed for expert users, with a suite of scripts and procedures for downloading initial, boundary and forcing data and for model set up, once the user has selected a specific geographical domain. This documentation provides the basic instructions to customize and apply the MITgcm-BFM model to different areas of the Mediterranean Sea.
The relocatable forecasting system was developed by OGS and it is built upon the coupled hydrodynamic-biogeochemical model based on the MITgcm and BFM open-source codes (Figure 1), both of which keep up to the state-of-the-art in the scope of ocean modeling. It is nested into the CMEMS Analysis and Forecasting system, and it includes the whole operational chain for running the simulations on a daily basis.
The general documentation for the coupled (hydrodynamic-biogeochemical) model MITgcm-BFM can be found at the following links:
- hydrodynamic model: http://mitgcm.org
- biogeochemical model: https://cmcc-foundation.github.io/ - www.bfm-community.eu/
- coupler (MITgcm-BFM): https://gmd.copernicus.org/articles/10/1423/2017/
The “relocability” means that an experienced user should be able to implement and run the forecasting system on his/her own HPC platform without too many efforts. Indeed, the operational chain includes pre-processing, launch and post-processing steps, i.e.:
- selection, “cut” and interpolation of the initial (ICs) and open boundary conditions (OBCs) from CMEMS;
- integration of river contributions into the open boundary condition (OBCs: sections/”slices” derived from CMEMS);
- definition of surface and bottom biogeochemical fluxes;
- creation of model namelists containing all the runtime parameters;
- various scripts in order to manipulate meteorological forcings originating from different sources/services.
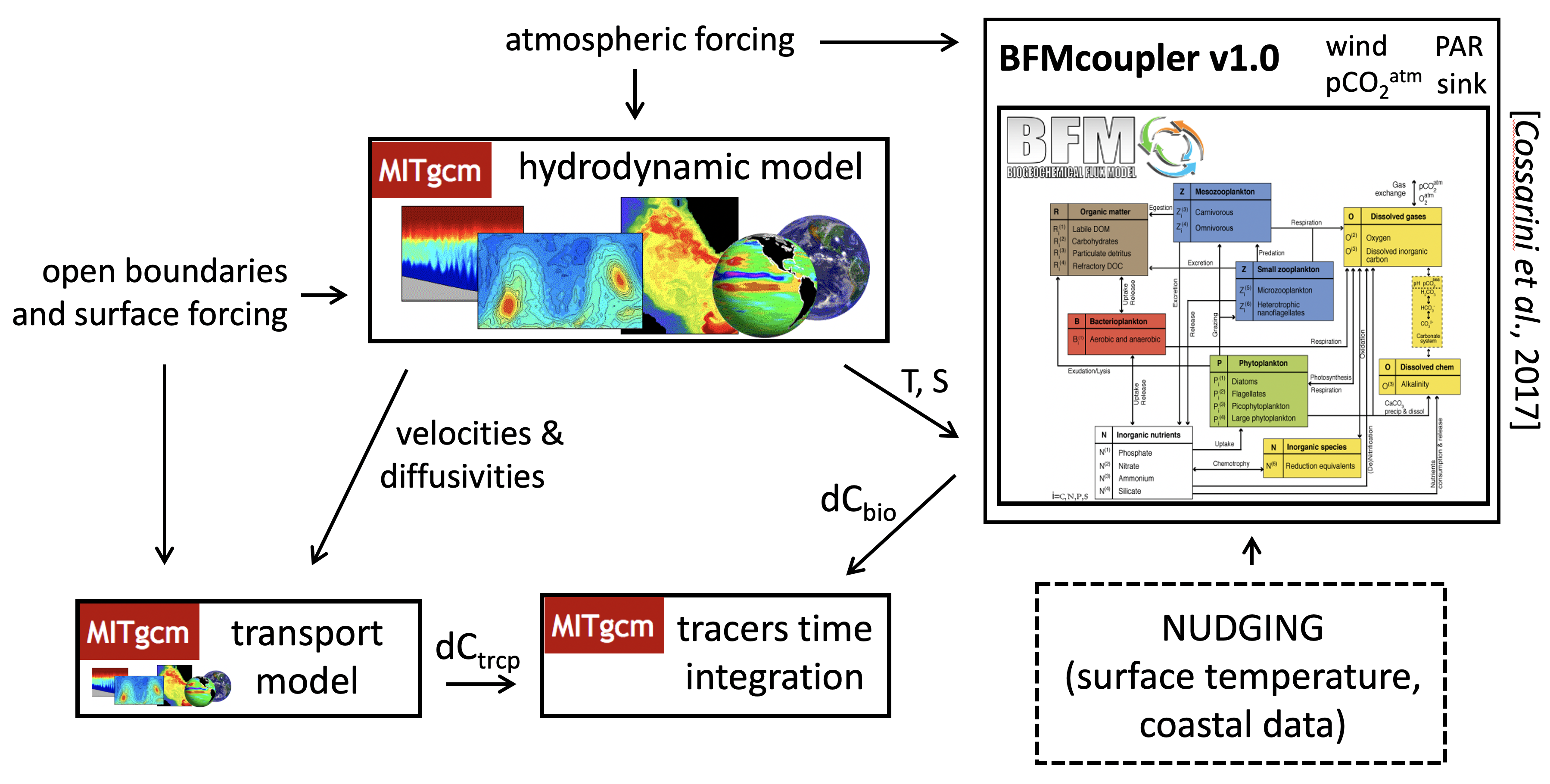
Figure 1 - Description of the MITgcm-BFM coupling used in SHAREMED.
The forecasting system was initially developed by OGS on the northern Adriatic Sea (at 1/128° resolution) and it is running operationally since several years, providing 72-hour forecasts (http://medeaf.ogs.it/adriatic). The implementation of the relocatable system relies on code sharing through GitHub repositories and, during the project, different customized configurations were developed at each study site (Figure 2).
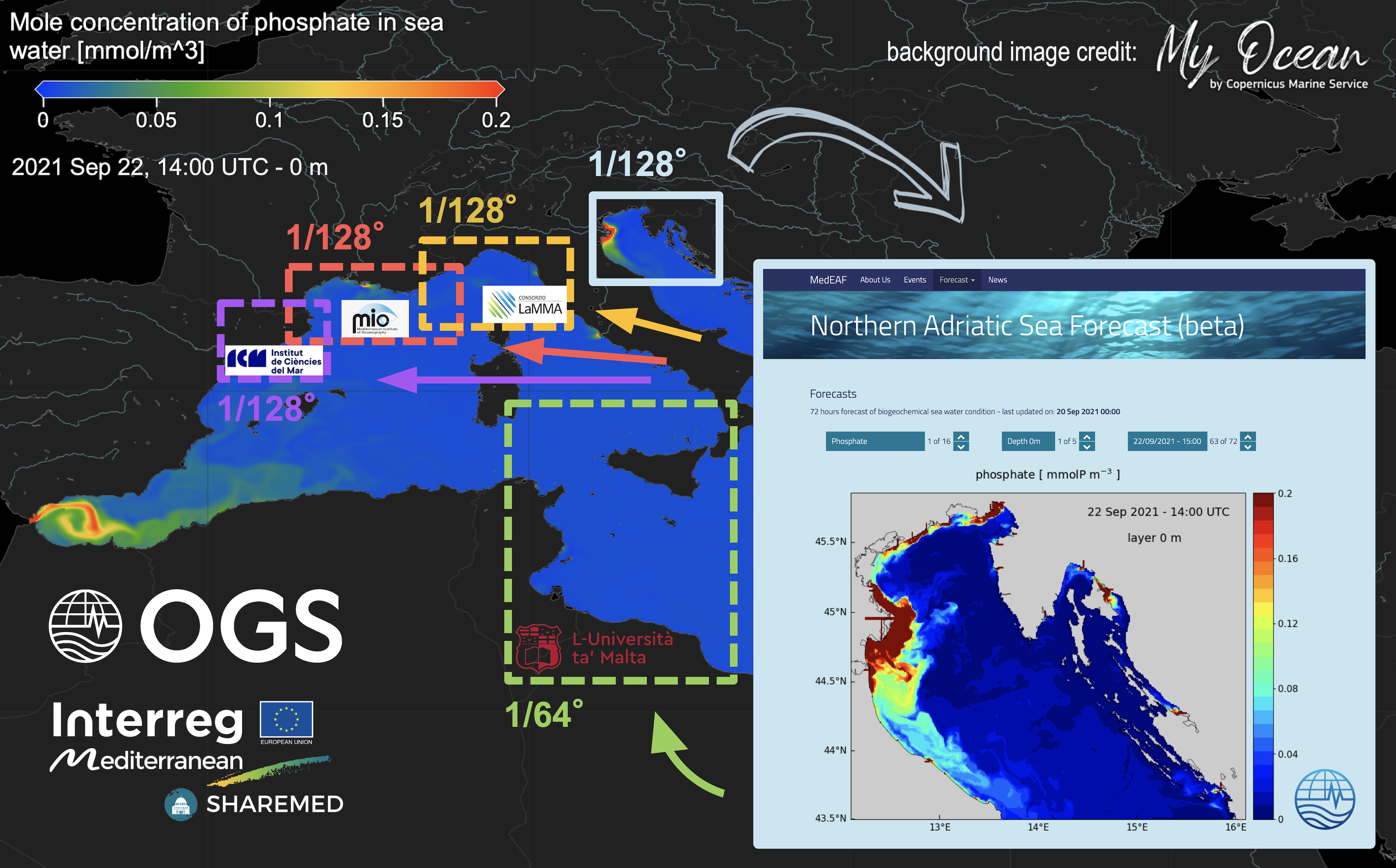
Figure 2 - The different configurations of the relocatable forecasting system in SHAREMED.
In order to build the MITgcm-BFM executable, the user must follow the steps below.
Define your ModelBuild directory and clone from the GitHub repository the tool for building the model:
DIR=ModelBuild
git clone git@github.com:inogs/MITgcmBFM-build.git $DIR
Download the source code of BFM, of the coupler and of MITgcm:
cd $DIR
git checkout main
./downloader_MITgcm_bfm.sh
Edit builder_MITgcm_bfm.sh, to set debug mode or not, compiler, modules, etc…
For example:
MIT_COMPILER=intel (gfortran)
export MODULEFILE=$PWD/compilers/machine_modules/g100.intel (fluxus.gfortran, yourmachine.yourcompiler)
Put your HPC-platform-specific INC_FILE for BFM in:
$BFMDIR/compilers/$INC_FILE (e.g., x86_64.LINUX.gfortran.inc)
Compile and build BFM:
./builder_MITgcm_bfm.sh -o bfm
Decide the number of cores and the domain decomposition of your model run, configuring your SIZE.h (e.g., choosing among the various options of the northern Adriatic benchmark experiment). E.g., for 95 cores:
cp presets/NORTH_ADRIATIC/SIZE.h_095p presets/NORTH_ADRIATIC/SIZE.h
Configure/customize the setup:
./configure_MITgcm_bfm.sh
If needed, update the MYCODE directory with your own modified routines (different OBCs, meteorological forcing files, etc...).
Put your HPC-platform-specific INC_FILE for MITgcm in:
$PWD/compilers/$INC_FILE (e.g., x86_64.LINUX.gfortran.inc).
Compile and build MITgcm: use the same builder used for compiling BFM (i.e., builder_MITgcm_bfm.sh)
./builder_MITgcm_bfm.sh -o MITgcm
The executable (mitgcmuv) is in $PWD/MITGCM_BUILD.
In this section we describe the main dependencies in the code pipeline and we provide a primer for the production workflow, from the download of the code to the installation of the chain.
Clone the operational chain from the GitHub repository.
git clone git@github.com:inogs/MITgcm_BFM_chain.git
The installation of the chain relies on the script called mit_setup_chain.ksh. In order to run it, the user must set properly the mit_profile__machine.src_inc, where machine is the name of the host, as set up in the MIT_HOSTNAME variable.
With this configuration script, the user must set some variables and run some commands, depending on the machine used (i.e., the available computing facility). The structure of this file is explained below, so that the user/installer can configure it without difficulties.
-
MIT_NETCDF_PATH: is the path of folder containing the NetCDF library on the machine (one can locate easily, for instance, loading the proper module, and spotting the folder in theLD_LIBRARY_PATHenvvariable); -
MIT_DATE_PATH: the path ofdatecommand (almost certainly will be /usr/bin/date); -
MIT_NCKS_PATH: the path ofnckson the machine; -
MIT_NCFTPGET_PATH: the path ofncftpgeton the machine; -
MITGCMUV_PATH: the path of the executable produced during the compilation of the model (see section above). This will be located in theMITGCM_BUILDsubfolder of the model folder; -
MOTUCLIENT_UNPACK_DIR: a path where the toolmotuclientwill be unpacked (usually set to$MIT_BINDIR/motuclient); -
PYTHON3_PATH: the path of a validpython3interpreter. It is used just to create virtual environment, the interpreter included in this environment will be used.
-
mit_mkdir <dir>: creates the directory<dir>with all its parents and checks for the success; -
symlink_if_not_present <path> <link>: symbolic links the<path>to<link>if<link>is not already present. If it is so, the function does nothing. If<path>it not present, it returns an error state; -
clone_or_update <git_link> <path> <branch>: if<path>is not present, it clones the<git_link>repo to<path>checking out branch<branch>(notice: if<path>is present it does nothing, the 'update' part has still to be implemented); -
download_and_unpack <url> <path> <filename> <extraction_dir>: downloads file at<url>placing it to<path>/<filename>. Then it creates the directory<extraction_dir>and extracts the contents to it. If some step fail, the function returns an error status; -
set_python_environment <pyhton3_path> <requirements>: uses<python3_path>to create an environment according to the variable$MIT_VENV_1(set inmit_profile.src_inc). Then it activates the environment, and usespipto install all the requirements specified in<requirements>(thepython3path is set for convenience in the variablePYTHON3_PATH. The requirements are listed in the file$MIT_HOME/requirements.txt).
Note that all the functions above are idempotent up to an upgrade coming from online repositories. This means that a script containing these function can be safely executed n times without spoiling the setup.
Although the exact command depends on the specific machine (i.e., modules to load, particular env variable to set) the structure of the file is the following:
# env variable section
mit_batch_prex "export LD_LIBRARY_PATH=${MIT_LIBDIR}:${MIT_NETCDF_PATH}:${LD_LIBRARY_PATH}"
mit_batch_prex "mit_mkdir $MIT_BINDIR"
# symlink section
mit_batch_prex "symlink_if_not_present $MIT_DATE_PATH $MIT_BINDIR/date"
mit_batch_prex "symlink_if_not_present $MIT_NCKS_PATH $MIT_BINDIR/ncks"
mit_batch_prex "symlink_if_not_present $MIT_NCFTPGET_PATH $MIT_BINDIR/ncftpget"
mit_batch_prex "symlink_if_not_present $MITGCMUV_PATH $MIT_BINDIR/mitgcmuv"
# git section
mit_batch_prex "clone_or_update $MIT_BITSEA_REPO $MIT_BITSEA $MIT_BITSEA_BRANCH"
mit_batch_prex "clone_or_update $MIT_POSTPROC_REPO $MIT_POSTPROCDIR $MIT_POSTPROC_BRANCH"
mit_batch_prex "clone_or_update $MIT_BC_IC_REPO $MIT_BC_IC_FROM_OGSTM_DIR $MIT_BC_IC_BRANCH"
# motuclient section
mit_batch_prex "download_and_unpack $MIT_MOTUCLIENT_URL $MIT_BINDIR motuclient.tar.gz $MOTUCLIENT_UNPACK_DIR"
mit_batch_prex "symlink_if_not_present $MOTUCLIENT_UNPACK_DIR/src/python $MIT_BINDIR/motuclient-python"
# python3 section
mit_batch_prex "set_python_environment $PYTHON3_PATH $MIT_HOME/requirements.txt"
# module section
# ad libitum
After setting mit_profile__machine.src_inc, create a script (e.g., chain_env.sh) with the following content (export and alias).
NOTE: given the example below, you could put it in /yourpath/.
-------------------------------------------------------
# MIT_HOME is your MITgcm_BFM_chain directory, the root of the git repository
export MIT_HOME=/yourpath/MITgcm_BFM_chain
# your machine name
export MIT_HOSTNAME=fluxus
# your work-launch (e.g., "scratch" area) directory in a parallel filesystem
export MIT_WORKDIR=/yourpath/WORK
export MIT_VERSION_NUMBER=1
export MIT_STAGE=devel
alias mitcd="cd $MIT_HOME ; pwd"
-------------------------------------------------------
Then do:
source chain_env.sh
cd $MIT_HOME/bin/src
For a first check that everything works, compile and install all the scripts:
make
make install
Add a line referring to your mit_profile__<yourmachinename>.inc in the SCRIPTS list of the Makefile ($MIT_HOME/bin/src/Makefile) and perform another (updated) installation:
make
make install
Once the job scheduler options are set (consult your IT manager for these details), you can launch your model run (with bulletin date: 19/10/2021, as an example).
cd $MIT_HOME/bin
./mit_start.ksh --pass --job-multiple --force --try-resume --without-kill --rundate 20211019
mit_start.ksh generates the log directory, where the single jobs and/or the single executables can be submitted or launched one by one for each of the three “stages” of a forecast run: preprocessing, model run, postprocessing.
To conclude, the compilation phase will produce an executable (MITgcm-BFM) called mitgcmuv. For running the model:
mit_start.ksh should run the whole model chain (if there are no setup/runtime errors). The job produced by mit_start.ksh (which needs the machine-specific scheduler options) will look for the MITgcm-BFM executable located in $MIT_HOME/HOST/$MIT_HOSTNAME/bin/ (together with the other executables: date, ncks...).
The executable can be renamed "mitgcmuv_<number_of_cores>", where the number of cores is specified in mit_mpi__intelmpi.src_inc.
copy the mitgcmuv executable in your run directory and run the simulation using a customized job for your local scheduler.
Model output is dumped in the native MITgcm format, in png images or in NetCDF products, organized in the same way as the CMS modelling products for the Mediterranean Sea. Figure 3 shows an example of model output published on a dedicated web page (https://medeaf.ogs.it/adriatic).
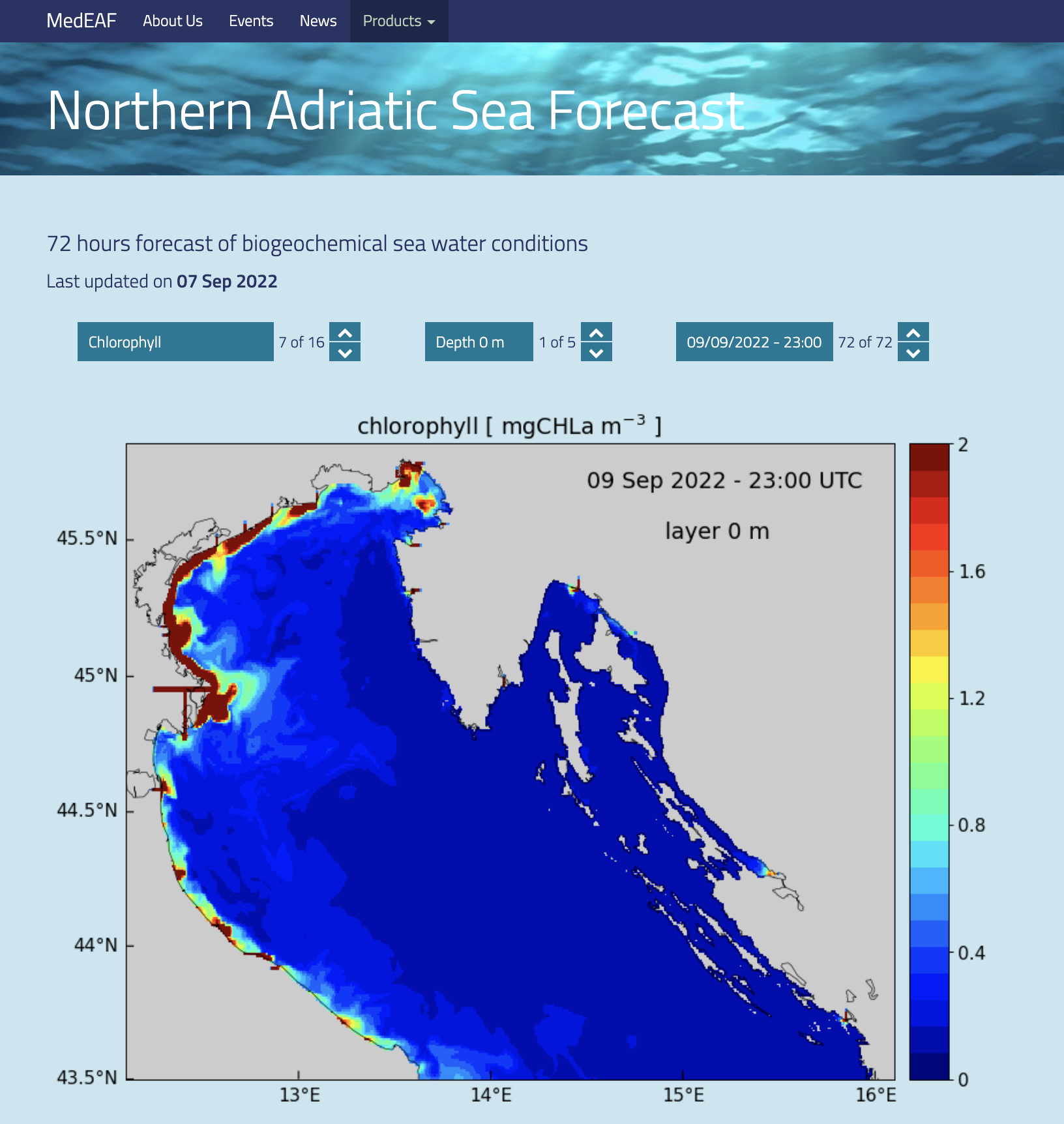
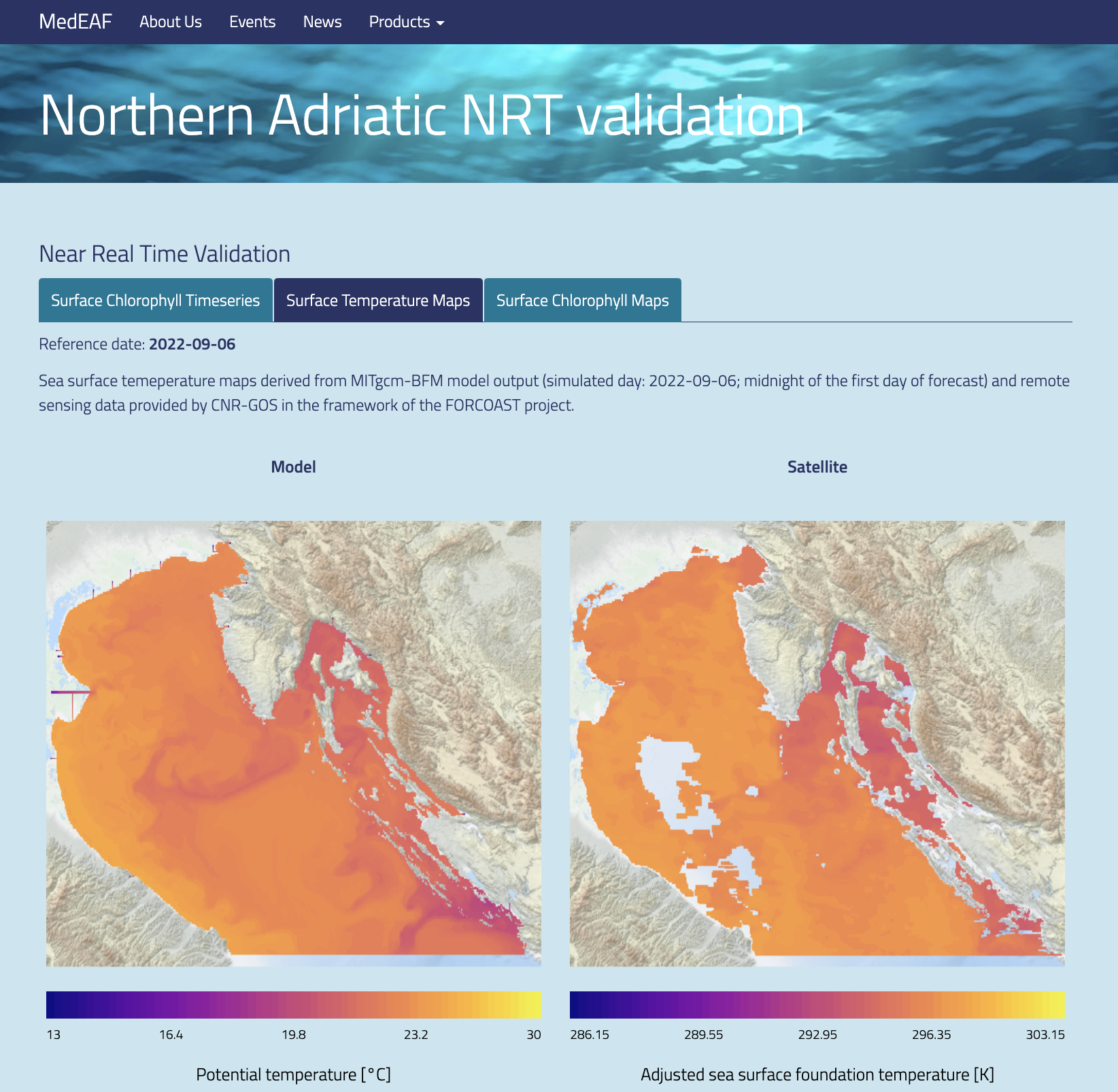
Figure 3 - Examples of model output published on dedicated web pages: chlorophyll forecast fields (left) and SST comparisons with satellite data (right).
NetCDF products can be also stored on specific web services, like the THREDDS server shown in Figure 4.
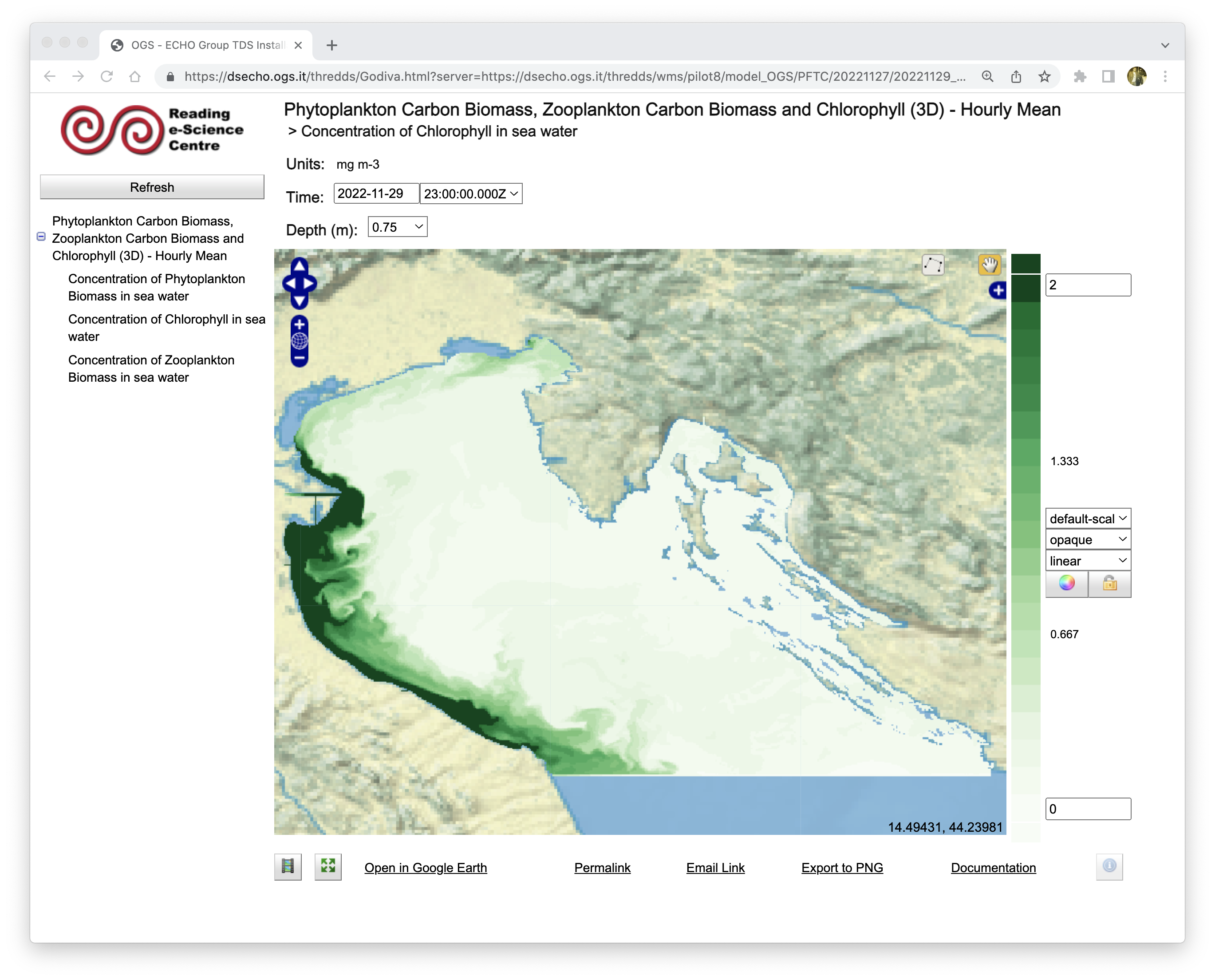
Figure 4 - Example of model output stored on a dedicated THREDDS server and visible through the “preview” tools available.
Campin, J.-M.; Heimbach, P.; Losch, M.; Forget, G.; edhill3; Adcroft, A.; amolod; Menemenlis, D.; dfer22; Hill, C.; et al. MITgcm/MITgcm: mid 2020 version; Zenodo, 2020
Cossarini, G.; Querin, S.; Solidoro, C.; Sannino, G.; Lazzari, P.; Di Biagio, V.; Bolzon, G. Development of BFMCOUPLER (v1.0), the coupling scheme that links the MITgcm and BFM models for ocean biogeochemistry simulations. Geosci. Model Dev. 2017, 10, 1423–1445, doi:10.5194/gmd-10-1423-2017
Marshall, J.; Adcroft, A.; Hill, C.; Perelman, L.; Heisey, C. A finite-volume, incompressible Navier Stokes model for studies of the ocean on parallel computers. J. Geophys. Res. Oceans 1997, 102, 5753–5766, doi:10.1029/96JC02775
Silvestri, C.; Bruschi, A.; Calace, N.; Cossarini, G.; Angelis, R.D.; Biagio, V.D.; Giua, N.; Saccomandi, F.; Peleggi, M.; Querin, S.; et al. CADEAU project - final report. 2020, 4399279 Bytes, doi:10.6084/M9.FIGSHARE.12666905.V1
Vichi, M.; Lovato, T.; Lazzari, P.; Cossarini, G.; Gutierrez Mlot, E.; Mattia, G.; Masina, S.; McKiver, W.J.; Pinardi, N.; Solidoro, C.; et al. The Biogeochemical Flux Model (BFM): Equation Description and User Manual. BFM version 5.1; BFM Consortium, 2015
test