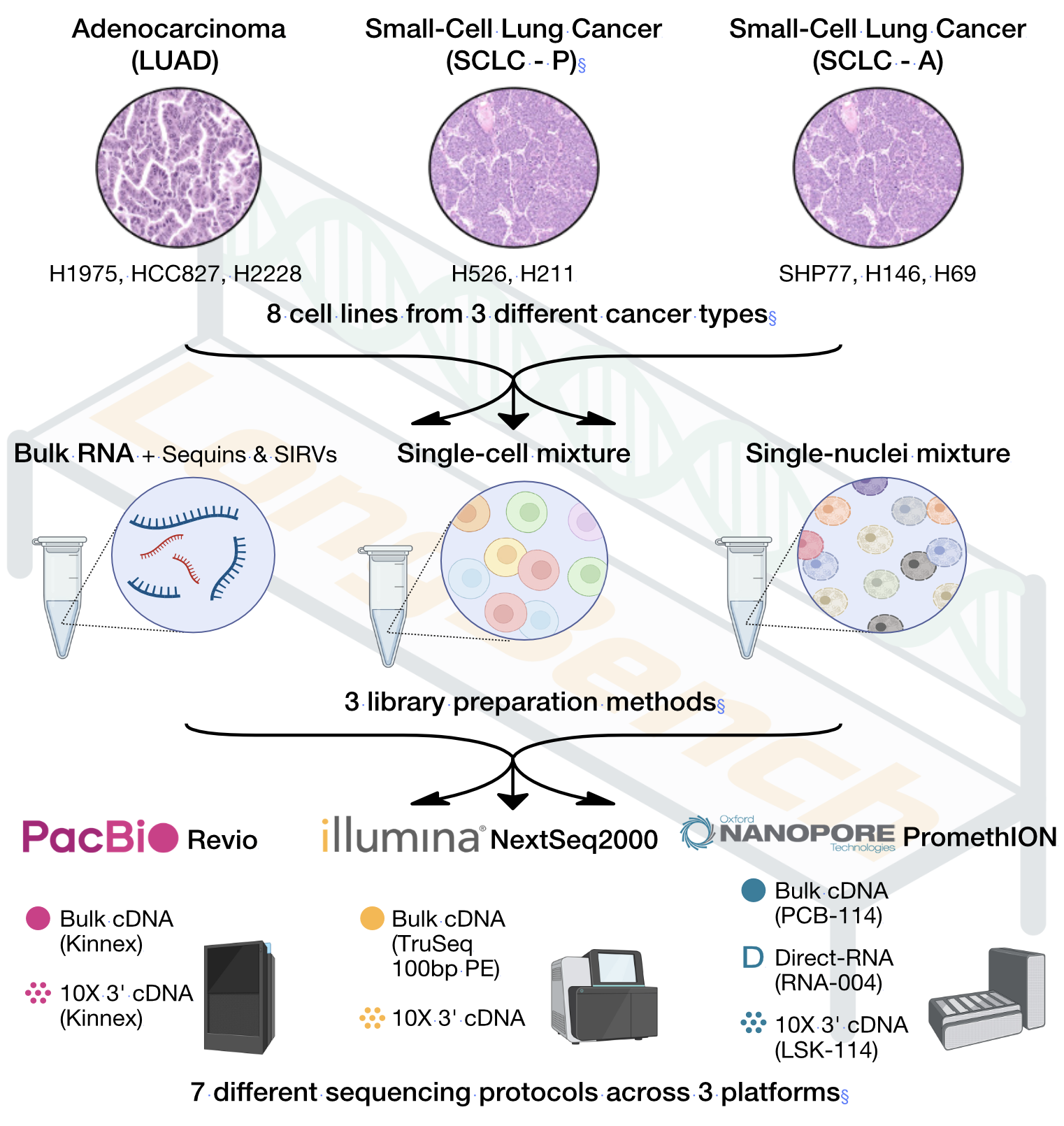
LongBench: a multi-platform reference dataset spanning bulk, single-cell, and single-nucleus transcriptomics across eight human lung cancer cell lines with synthetic spike-in controls.
LongBench is a comprehensive benchmarking dataset designed to fill these critical gaps. Derived from eight lung cancer cell lines with synthetic RNA spike-ins, LongBench includes bulk, single-cell, and single-nucleus RNA-seq data from three state-of-the-art long-read sequencing platforms — ONT PCR-cDNA, ONT direct RNA, and PacBio Kinnex — alongside Illumina short-read data for robust cross-platform comparisons.
The LongBench dataset is a valuable resource for benchmarking and improving sequencing protocols and bioinformatics tools. With the LongBench dataset we present a systematic evaluation of transcript capture, quantification, and differential expression analyses, examining the strengths and limitations of each sequencing platform in various biological contexts, enabling researchers to make more informed decisions on platform and method selection.
More details of the dataset can be found here.
Some example applications of the LongBench dataset include:
- Studying biological questions including isoform diversity, variant calling, allele-specific expression, and RNA modifications across lung cancer subtypes.
- Benchmarking analysis tools and method development for isoform quantification, differential expression (DE), differential transcript usage (DTU), and variant calling.
- Cross-platform comparisons of accuracy, sensitivity, and biases between ONT, PacBio, and Illumina sequencing technologies.
The LongBench dataset is publicly hosted on AWS S3 and includes:
- raw FASTQ – bulk, single-cell, and single-nucleus RNA-seq
- POD5 files – ONT direct-RNA reads for RNA-modification analysis
- Count matrices - processed gene/transcript expression tables
- Cell line annotation - metadata for single-cell and single-nucleus datasets
Install the AWS CLI:
pip install awsclilist data in the s3 bucket
aws s3 ls s3://longbench-data/ --no-sign-requestDownload data
aws s3 sync s3://longbench-data/<dir name> <local dir name> --no-sign-requestThe following tutorials are available that demonstrate how to analyse the LongBench data:
- Bulk RNA-seq Differential analysis with edgeR
- Single-cell RNA-seq analysis with Seurat (Coming soon)
If you use the LongBench dataset, please cite:
Yupei You, Ashleigh Solano, James Lancaster, Margaux David, Changqing Wang, Shian Su, Kathleen Zeglinski, Reza Ghamsari, Manveer Chauhan, Josie Gleeson, Yair D. J. Prawer, Jin Ng, Benedicte Dubois, Isabelle Cleynen, Marie-Liesse Asselin-Labat, Kate D. Sutherland, Michael B. Clark, Quentin Gouil, Matthew E. Ritchie. Benchmarking long-read RNA-sequencing technologies with LongBench: a cross-platform reference dataset profiling cancer cell lines with bulk and single-cell approaches. bioRxiv 2025.09.11.675724. https://doi.org/10.1101/2025.09.11.675724